Continuous biomolecular fractionation (summary) - Micro/Nanofluidic BioMEMS Group
Executive Summary
Biomolecule analysis is often compared to finding “a needle in a haystack”, so the importance of biosample fractionation cannot be exaggerated. Traditionally, Gel electrophoresis, gel-exclusion chromatography and other filtration techniques have been used for bioseparation. In addition to well-known drawbacks such as manual operation, slow separation rate, and need for large equipments, the science behind the molecular sieving and filtration is still yet to be fully clarified. One issue is that most molecular sieves (gels) are random nanoporous materials, making it difficult to control / optimize the separation process. In our group, patterned regular sieving structures and nanofilters have been sought as an alternative to conventional separation method: recent developments in micro nanofluidic sieves and filters have demonstrated superior performance for both analytical and preparative separation of various physiologically relevant macromolecules, including proteins.
Bow, H., Pivkin, I., Diez-Silva, M., Goldfless, S.J., Dao, M., Niles, J.C., Suresh, S. & Han, J. “A microfabricated deformability-based flow cytometer with application to malaria,”Lab on a Chip, 2011, 11(6), 1065-1073
Malaria resulting from Plasmodium falciparum infection is a major cause of human suffering and mortality. Red blood cell (RBC) deformability plays a major role in the pathogenesis of malaria. Here we introduce an automated microfabricated “deformability cytometer” that measures dynamic mechanical responses of 103 to 104 individual RBCs in a cell population. Fluorescence measurements of each RBC are simultaneously acquired, resulting in a population-based correlation between biochemical properties, such as cell surface markers, and dynamic mechanical deformability. This device is especially applicable to heterogeneous cell populations. We demonstrate its ability to mechanically characterize a small number of P. falciparum-infected (ring stage) RBCs in a large population of uninfected RBCs. Furthermore, we are able to infer quantitative mechanical properties of individual RBCs from the observed dynamic behavior through a dissipative particle dynamics (DPD) model. These methods collectively provide a systematic approach to characterize the biomechanical properties of cells in a high-throughput manner.
Sung Jae Kim, Sung Hee Ko, Kwan Hyoung Kang and Jongyoon Han, “Direct Seawater Desalination by Ion Concentration Polarization,” Nature Nanotechnology, 2010, 5, 297-301.
A shortage of fresh water is one of the acute challenges facing the world today. An energy-efficient approach to converting sea water into fresh water could be of substantial benefit, but current desalination methods require high power consumption and operating costs or large-scale infrastructures, which make them difficult to implement in resource-limited settings or in disaster scenarios. Here, we report a process for converting sea water (salinity ~500 mM or ~30,000 mg/L) to fresh water (salinity <10 mM or <600 mg/L) in which a continuous stream of sea water is divided into desalted and concentrated streams by ion concentration polarization, a phenomenon that occurs when an ion current is passed through ion-selective membranes. During operation, both salts and larger particles (cells, viruses and microorganisms) are pushed away from the membrane (a nanochannel or nanoporous membrane), which significantly reduces the possibility of membrane fouling and salt accumulation, thus avoiding two problems that plague other membrane filtration methods. To implement this approach, a simple microfluidic device was fabricated and shown to be capable of continuous desalination of sea water (~99% salt rejection at 50% recovery rate) at a power consumption of less than 3.5 Wh/L, which is comparable to current state-of-the-art systems. Rather than competing with larger desalination plants, the method could be used to make small- or medium-scale systems, with the possibility of battery-powered operation. (detail)
Yong-Ak Song, Michael Chan, Chris Celio, Steven R. Tannenbaum, John S. Wishnok, and Jongyoon Han, “Free-Flow Zone Electrophoresis of Peptides and Proteins in PDMS Microchip for Narrow pI Range Sample Frefractionation in Mass Spectrometry,” Analytical Chemistry, 2010, 82, 2317-2325.
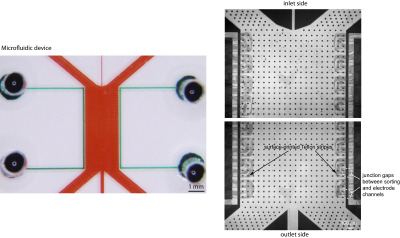
In this paper, we are evaluating the strategy of sorting peptides / proteins based on the charge to mass ratio without resorting to ampholytes and / or isoelectric focusing, using a single- and two-step free-flow zone electrophoresis. We developed a simple fabrication method to create a salt bridge for free-flow zone electrophoresis in PDMS chips by surface printing a hydrophobic layer on a glass substrate. Since the surface-printed hydrophobic layer prevents plasma bonding between the PDMS chip and the substrate, an electrical junction gap can be created for free-flow zone electrophoresis. With this device, we demonstrated a separation of positive and negative peptides and proteins at a given pH in standard buffer systems, and validated the sorting result with LC/MS. Furthermore, we coupled two sorting steps via off-chip titration, and isolated peptides within specific pI ranges from sample mixtures, where the pI range was simply set by the pH values of the buffer solutions. This free-flow zone electrophoresis sorting device, with its simplicity of fabrication, and a sorting resolution of 0.5 pH unit, can potentially be a high-throughput sample fractionation tool for targeted proteomics using LC/MS.
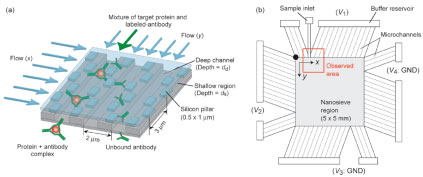
Yamada, M., Mao, P., Fu, J. and Han, J., “Rapid Quantification of Disease-Marker Proteins Using Continuous-Flow Immunoseparation in a Nanosieve Fluidic Device,” Analytical Chemistry, 81, 7067-7074 (2009).
This paper reported the rapid and quantitative immunoseparation assay of proteins by utilizing the anistropically patterned nanosieve array (ANA) structures. Human C-reactive protein (CRP) and human chorionic gonadotropin (hCG) are examined using fluorescent-labeled polyclonal antibodies. The size of the immunocomplex and the field strength are critical for separation. The application of the ANA structure for studying the binding kinetics of protein and antibody is also demonstrated.
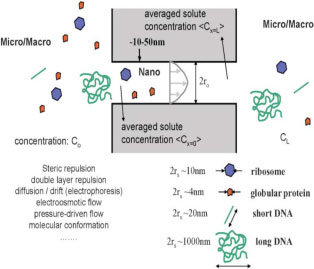
Jongyoon Han, Jianping Fu, and Reto B. Schoch, “Molecular Sieving Using Nanofilters: Past, Present and Future,” Lab on a Chip, 7, 23-33, 2007
This paper reviewed relevant theoretical developments in the hindered transport theory and summarized recent (1990s and after) developments of artificial molecular sieving systems both from a scientific and engineering point of view.
- Zi Rui Li, G. R. Liu, J. Han, Y. Cheng, Y. Z. Chen, J.-S. Wang, N. G. Hadjiconstantinou, “Analytical description of Ogston-regime biomolecule separation using nanofilters and nanopores” Physical Review E 2009, accepted for publication.
- Li, Z.R., Liu, G.R., Han, J., Chen, Y.Z., Wang, J.H. and Hadjiconstantinou, N.G., “Transport of biomolecules in asymmetric nanofilter arrays,” Analytical and Bioanalytical Chemistry, 394, 427-435 (2009).
- Zi Rui Li, Gui Rong Liu, Yu Zong Chen, Jian-Sheng Wang, Hansen Bow, Yuan Cheng, Jongyoon Han, “Continuum Transport Model of Ogston Sieving in Patterned Nanofilter Arrays for Rod-like Biomolecule Separation,” Electrophoresis, 29, 329–339. 2008.**
In order to optimize nanofluidic biomolecule separation systems, development of theoretical model is desirable. For the case of rigid, rod-like biopolymers (such as short, double-stranded DNA), the Ogston sieving process can be modeled by considering the rotational degree of freedom of rod-like molecules near the wall. Using this simple idea, we have developed a continuum model for Ogston sieving process, by defining an “entropic driving force” term in the master transport equation. Since the calculation of this model is relatively fast, it is useful for repeated modeling of various device structures for the optimization of the separation system. Extension of this idea to the macrotransport theory will allow one to generate an analytical expression for sieving resolution, which would be instrumental in gaining important physical intuition on the process of molecular sieving in nanochannels.
- Duong-Hong, D., J. Han, J.-S. Wang, N. G. Hadjiconstantinou, Y. Z. Chen, and G.-R. Liu, “Realistic Simulations of Combined DNA Electrophoretic and Electroosmotic Flows in Nano-Fluidic Devices,” Electrophoresis, 29, 4880-4886, 2008.
- Duc Duong-Hong, Jian-Sheng Wang, G. R. Liu, Yu Zong Chen, Jongyoon Han, G. Hadjiconstantinou, “Dissipative particle dynamics simulations of electroosmotic flow in nano-fluidic devices,” Microfluidics and Nanofluidics, 4(3), 219-225, 2007
Entropic trapping of long, flexible polymer (Han et al. 2000, Science) is hard to model accurately, due to complexities in polymer conformation as well as local electroosmotic flow generated whenever a polyelectrolyte is trapped via non-electrical means. While stochastic simulation techniques are required, full blown molecular dynamics is too slow to model such a mesoscale problem. In these two publications, we have developed a Diffuse Particle Dynamics (DPD) simulation model as a “coarse-grained MD” technique to simulate the complicated, multiscale problem of entropic trapping in its full detail. In this model, one can simulate both stochastic polymer dynamics and hydrodynamic interaction of DNA polymer with surroundings. As a result, so-called free-draining electrophoresis behavior (length-independent mobility) and Zimm-model diffusion (length-dependent diffusivity) of DNA are correctly modeled. This will allow modeling both molecular sieving and dispersion, an important step toward engineering model for optimization of separation systems.
- Bow, H., Fu, J. and Han, J., “Decreasing effective nanofilter size by modulating electrical double layers: Separation enhancement in microfabricated nanofilters,” Electrophoresis, 29, 1-6 (2008).
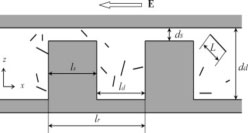
This work investigated methods that can be used in the sieves to increase separation selectivity and resolution. Selectivity achieved in devices with different dimensions is compared. The electrostatic repulsion between the charged molecules and the charged nanofluidic structure is also exploited. Decreasing buffer ionic strength in the nanofilter array led to higher selectivity and separation resolution for 100-1000bp DNA.
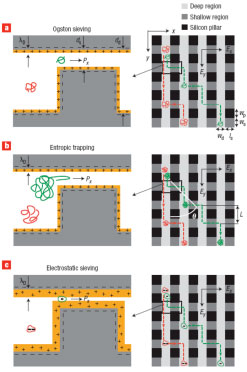
Fu, J., Schoch, R. B., Stevens, A. L., Tannenbaum, S. R. and Han, J. “A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins,” Nature Nanotechnology 2, 121-128 (2007).
This paper reported a microfabricated anisotropic sieving structure consisting of a two-dimensional periodic nanofluidic filter array. The designed structural anisotropy causes different-sized or -charged biomolecules to follow distinct trajectories, with either Ogston sieving, entropic trapping or electrostatic sieving, leading to efficient separation. Continuous-flow size based separation of DNA and proteins, as well as electrostatic separation of proteins, were achieved in just a few minutes.
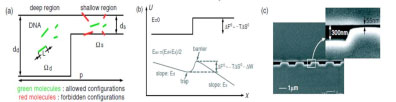
Fu, J., Yoo, J. and Han, J. “Molecular sieving in periodic free-energy landscapes created by patterned nanofilter arrays,” Physical Review Letters 97, 018103.1-4 (2006).
By using a theoretical model based on equilibrium partitioning theory, the Ogston-like sieving process of rodlike DNA in patterned periodic nanofluidic filter arrays is studied. The electrophoretic motion of DNA through the array is described as a biased Brownian motion overcoming periodically modulated free-energy landscape. Experimental evidence shows the crossover from Ogston-like sieving to entropic trapping, depending on the ratio between nanofilter constriction size and DNA size.
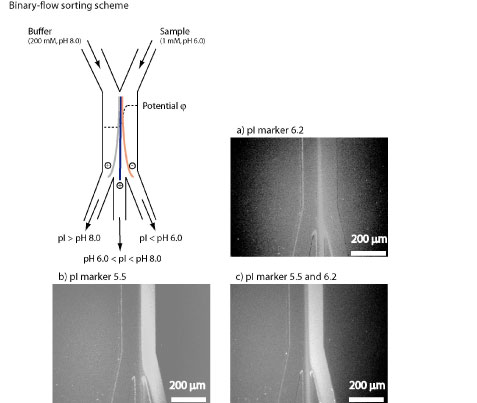
Song, Y.-A., Hsu, S., Stevens, A. and Han, J. “Continuous-flow pI-based sorting of proteins and peptides in a microfluidic chip using diffusion potential,” Analytical Chemistry 78, 3528-3536 (2006).
This paper reported a continuous-flow, ampholyte-free, isoelectric point (PI)-based sorting technique for proteins and peptides in a microfluidic chip. The electrophoretic field required to run the sorting is generated by the diffusion of buffer ions in situ, at the liquid junction between two laminar flows within the microfluidic channel. The sorting resolution could achieve ~0.1 pH unit and the relatively high flow rate enables high-throughput sample preparation.
Fu, J., Mao, P. and Han, J. “A nanofilter array chip for fast gel-free biomolecule separation,” Applied Physics Letters 87, 263902.1-3 (2005).
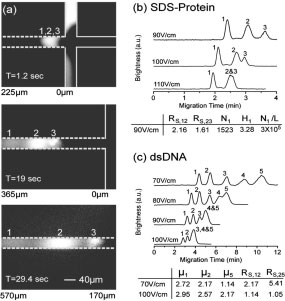
This paper reported size-separated sodium dodecyl sulfate (SDS)-protein complexes and small DNA molecules in nanofilter array chips based on the Ogston sieving mechanism. Nanofilter arrays with a gap size of 40- 180 nm were fabricated and characterized. Complete separation of SDS-protein complexes and small DNA molecules were achieved in several minutes with a separation length of 5mm.
Wang, Y.-C., Choi, M. H. & Han, J. “Two-dimensional protein separation with advanced sample and buffer isolation using microfluidic valves,” Analytical Chemistry, 76, 4426-4431 (2004).
Methods are described to achieve more efficient multidimensional protein separation in a microfluidic channel. The new methods couple isoelectric focusing (IEF) with high ionic strength electrophoretic separations by active microvalve control in a microchip. Chip based IEF with CE or CGE are successfully integrated and 2D protein separation on-chip is achieved in 20 minute.