“Multilayer block copolymer meshes by orthogonal self-assembly”, Amir Tavakkoli K. G., Samuel M. Nicaise, Karim R. Gadelrab, Alfredo Alexander-Katz, Caroline A. Ross & Karl K. Berggren. Nature Communications, (2016)
DOI: 10.1038/ncomms10518
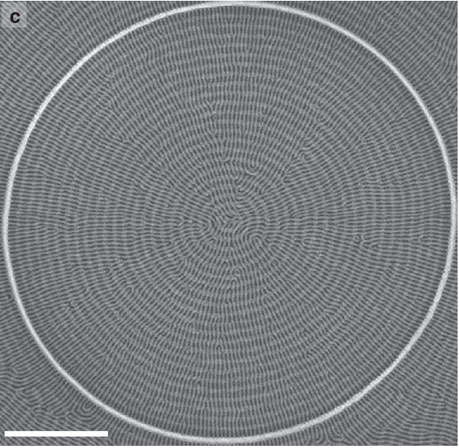
Continued scaling-down of lithographic-pattern feature sizes has brought templated self-assembly of block copolymers (BCPs) into the forefront of nanofabrication research. Technologies now exist that facilitate significant control over otherwise unorganized assembly of BCP microdomains to form both long-range and locally complex monolayer patterns. In contrast, the extension of this control into multilayers or 3D structures of BCP microdomains remains limited, despite the possible technological applications in next-generation devices. Here, we develop and analyse an orthogonal self-assembly method in which multiple layers of distinct-molecular-weight BCPs naturally produce nanomesh structures of cylindrical microdomains without requiring layer-by-layer alignment or high-resolution lithographic templating. The mechanisms for orthogonal self-assembly are investigated with both experiment and simulation, and we determine that the control over height and chemical preference of templates are critical process parameters. The method is employed to produce nanomeshes with the shapes of circles and Y-intersections, and is extended to produce three layers of orthogonally oriented cylinders.
Learn more about the researchers and their motivations behind this work in these articles from MIT news and Science News.
“The Orientations of Large Aspect-Ratio Coiled-Coil Proteins Attached to Gold Nanostructures“, Jae-Byum Chang, Yong Ho Kim, Evan Thompson, Young Hyun No, Nam Hyeong Kim, Jose Arrieta, Vitor R. Manfrinato, Amy E. Keating & Karl K. Berggren. Small (2016)
DOI: 10.1002/smll.201502419
Methods for patterning biomolecules on a substrate at the single molecule level have been studied as a route to sensors with single-molecular sensitivity or as a way to probe biological phenomena at the single-molecule level. However, the arrangement and orientation of single biomolecules on substrates has been less investigated. Here, the arrangement and orientation of two rod-like coiled-coil proteins, cortexillin and tropomyosin, around patterned gold nanostructures is examined. The high aspect ratio of the coiled coils makes it possible to study their orientations and to pursue a strategy of protein orientation via two-point attachment. The proteins are anchored to the surfaces using thiol groups, and the number of cysteine residues in tropomyosin is varied to test how this variation affects the structure and arrangement of the surface-attached proteins. Molecular dynamics studies are used to interpret the observed positional distributions. Based on initial studies of protein attachment to gold post structures, two 31-nm-long tropomyosin molecules are aligned between the two sidewalls of a trench with a width of 68 nm. Because the approach presented in this study uses one of twenty natural amino acids, this method provides a convenient way to pattern biomolecules on substrates using standard chemistry.